Single-Cell RNA-Sequencing Pipeline
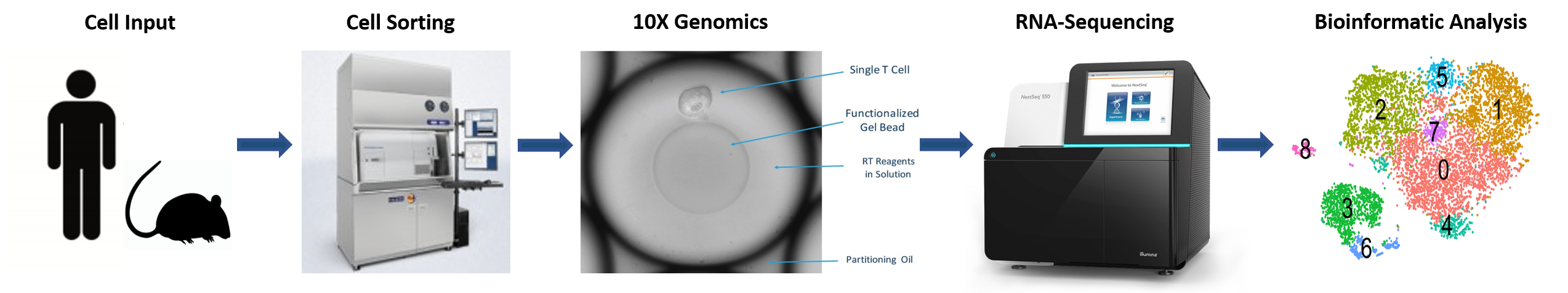
- EXPERIMENTAL DESIGN – The core offers an initial consultation to discuss sample preparation and experimental design
- CELL SORTING – Viable cell populations free of debris are isolated by flow cytometry
- 10X GENOMICS – Samples are run through 10X controller and libraries are generated
- RNA SEQUENCING – Pooled libraries are sequenced in partnership with the Molecular Biology Core Facilities (MBCF) at Dana-Farber Cancer Institute (DFCI)
- BIOINFORMATIC ANALYSIS – Raw data are processed using Cell Ranger and standard QC to generate count matrices and Loupe files.
Frequently Asked Questions
What's included in the price package?
Does the core provide cell hashing antibodies and or CITE-seq antibodies?
Yes, we can provide the relevant cell hashtag and CITE-seq antibodies, Fc blocking reagent, and staining buffer. If you wish to spike in custom antibodies, please order the antibody panels from Biolegend and send the relevant information to the core. After you book your experiment, please coordinate with us to pick up those reagents one day in advance.
Do I need to pool the cells from different populations myself if I plan to do Cell hashing?
Yes. You will need to count the cell number of each population, pool to the required proportions (usually in equal numbers) prior to submitting the pooled sample to us. We will then perform a confirmatory assessment of the cell concentration and viability of the sample prior to running on the 10x Controller.
Is sequencing cost included in the price package?
Yes.
What is included for data analysis?
Data generated from the core will be processed through the standard Cell Ranger pipeline (10X Genomics). Raw data, QC metric data, count matrices, and browsable data files in Loupe Cell format will be made available to investigators.
When will I expect to receive my data?
Our turnover time is generally around 10-12 weeks.
How do I prepare cells for Single-Cell Assays?
How many cells do I need to provide and how many cells can I expect to get information for?
We recommend loading between 20k live cells (for non-hashing experiments) and 60k live cells (for cell hashing experiments). Therefore, please provide at least double this cell number, so that there are enough cells to re-load the sample if a wetting failure occurs during the 10x encapsulation. The capture rate is approximately 80%, depending on cell type and cell quality.
How should the cells be prepared for 10X?
It is recommended that investigators should optimize their cell isolation procedure prior to 10X experiments. In general, we recommend cell viability of >85% for optimal cell input. For buffer recommendation please check here.
The Sorting Facility at CCP can be used to isolate viable cells or there are protocols available for cell isolation by MACs beads. For sorting, please check out our sample prep recommendation.
For extremely fragile cells, please check out this 10x recommendation document.
For single cell RNA-seq, we ask you to resuspend your cells in 0.4%BSA/PBS or 10x tested media.
For single nuclei RNA-seq, 1% BSA/PBS with 0.2U/μl RNase Inhibitor is required (users provide their own RNase Inhibitor). For ATAC-seq, we can provide the 20X Nuclei Buffer – please dilute 1:20 in nuclease-free water before use.
More on Single-Cell sample preparation, please check here.
What is your sequencing platform?
What is your sequencing platform?
Sequencing is performed by the Molecular Biology Core Facilities (MBCF) at Dana-Farber Cancer Institute (DFCI). We use Illumina NovaSeq 6000 (S4, S2, S1 and SP flowcell), HiSeq X, NextSeq 500 systems and NovaSeq X Plus.
Do you pool libraries for sequencing?
Yes. The pooled number depends on the cell input of the libraries and the outputs of sequencing platform.
How many reads per cell can I expect?
We aim to provide at least 20k reads/cell for gene expression libraries, as recommended by 10x Genomics. For Cite-seq libraries, we provide a sequencing depth of 5,000 reads/cell. If you are using more than 100 TotalSeq™ antibodies, we will increase the read number to 10,000 reads/cell. For cell hashing libraries, we provide a sequencing depth of 5,000 reads/cell.
Will I be able to get the raw sequencing data?
Yes, we routinely deliver BAM files, which FASTQ files (raw data) can be obtained from by performing bamtofastq function within Cell Ranger software.
What's the data output format?
What are the steps taken for QC?
The Cell Ranger pipeline performs initial QC and provides a matrix with counts per GEMs that are likely cells. We recommend performing extra-downstream filters based on number of genes per cell and mitochondrial content (appropriate thresholds may vary per experimental design and cell type).
How can I understand the data output and get recommendations of downstream analyses?
Please see our Data Output User Guide on the Resources & Disclaimers page.