Sequencing and Imaging Spatial Transcriptomic Services
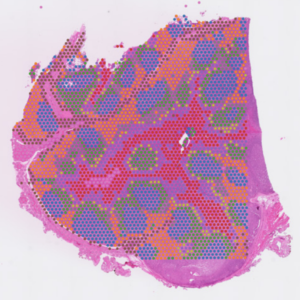
- Compatible with FFPE samples
- Analyze the whole transcriptome from the entire section (1-10 cell resolution)
- Discover new biomarkers, identify novel cell types and states
- Combined histological and gene expression data with easy to use software
- Slide preparation please refer to our Visium CytAssist and Visium HD slide preparation guide
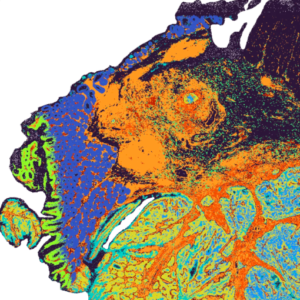
- Compatible with FFPE samples
- Data output is 2um x 2um barcoded area achieving single-cell spatial resolution
- Preview of Visium HD Poster and Public Visium HD data
- Slide preparation please refer to our Visium CytAssist and Visium HD slide preparation guide
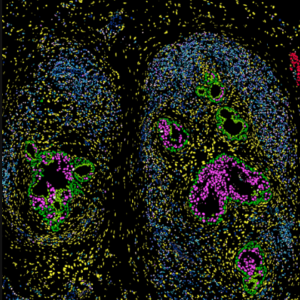
- Compatible with Fresh Frozen and FFPE samples
- Targeted gene expression information with subcellular resolution
- Characterize 100s to 1000s RNAs and multiplexed protein in cells
- Fast run time, pre-designed or customization of gene panels
- Slide preparation please refer to our Xenium slide preparation guide
- For Fresh Frozen tissues, please refer to Xenium Tissue Preparation Guide for FFT

Important Disclaimers
(Must read and accept prior to use of core)
Fluorescence in-situ hybridization and whole transcriptome spatial profiling experiments are cutting-edge technologies that offer exciting new research opportunities. Due to the high-risk nature of these technologies, we cannot guarantee a successful outcome with each experiment. We also cannot guarantee any specific metrics such as cells captures nor UMI’s per cell due to the high variability of these values across tissues, species, collection methods, processing protocols, and RNA quality.
Sample Preparation & Quality Disclaimer
As a core facility, we will be happy to provide relevant information on proper sectioning and staining techniques leading up to the use of the services offered to give the best chance at a desired outcome.
To prevent poor experimental outcomes, we require users to H&E stain each sample and submit the scanned images to the core. We ask that users inspect these images and the tissue structures that they want to characterize are present. We also highly encourage users to perform RNA extraction of tissue scrolls, and we offer to analyze these extracted transcripts on our Tape Station. We will also inspect the images and Tape Station results and notify the investigator prior to proceeding with the experiment of any indications of poor tissue quality.
Sample preparation and quality is of the utmost importance. Positive results from these two preliminary measures will not guarantee success, but they function to eliminate samples with a heightened risk of failure. At this point the investigator and PI will have the choice to proceed with risk or cancel the run.
In the event of PROCEEDING WITH RISK using a suboptimal sample, and a failure occurs, the investigator will be billed for reagents and labor costs associated with the experiment.
For Visium, there is a step in the library construction in which gives us an indication if the probes used were able to capture any RNA transcripts within the tissue. At this point, investigators can choose to proceed with sequencing. If the samples fail to pass this step and investigators choose to not proceed with sequencing, users will still be liable for reagents and labor costs.
Machine Disclaimer
Though small, there is always a risk for machine failure that is outside of our control. In this event, there is a chance that the samples may not be retrievable. We do not require back up slides in order to run users’ experiments, but we do encourage the use of blocks with sufficient tissue for multiple runs.
Data Storage Disclaimer
The Core promises to store user data for 90 days beginning on the day of file transfer. After these 90 days, the data will be deleted from our servers. Please communicate any difficulties viewing or accessing your data for we are not liable for any data issues past these 90 days.